Research
My
main research interest is in the area of statistical 3D curve shape
analysis of structures of RNAs and proteins. This research area is
multidisciplinary in that it involves statistics,
differential geometry, functional analysis, group algebra, computer
image analysis and bioinformatics.
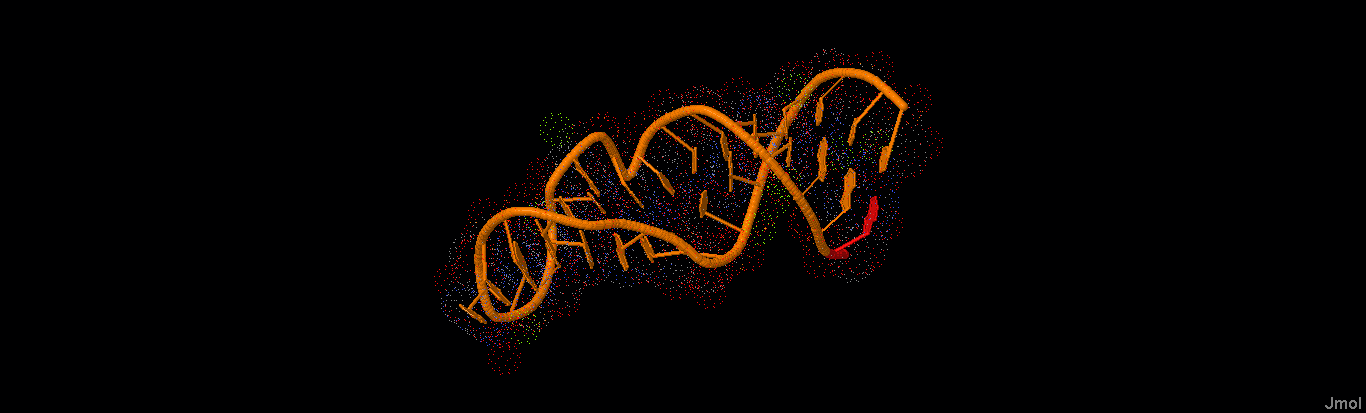
This work uses Elastic Shape Analysis (ESA), a mathematical framework which has been recently developed and used in computer vision, to provide new methodologies to perform comparison through a formal distance and classification of these biomolecules. ESA uses the geometry of the 3D structure of curves and surfaces to produce mathematically formal distances, geodesics and alignments between them.
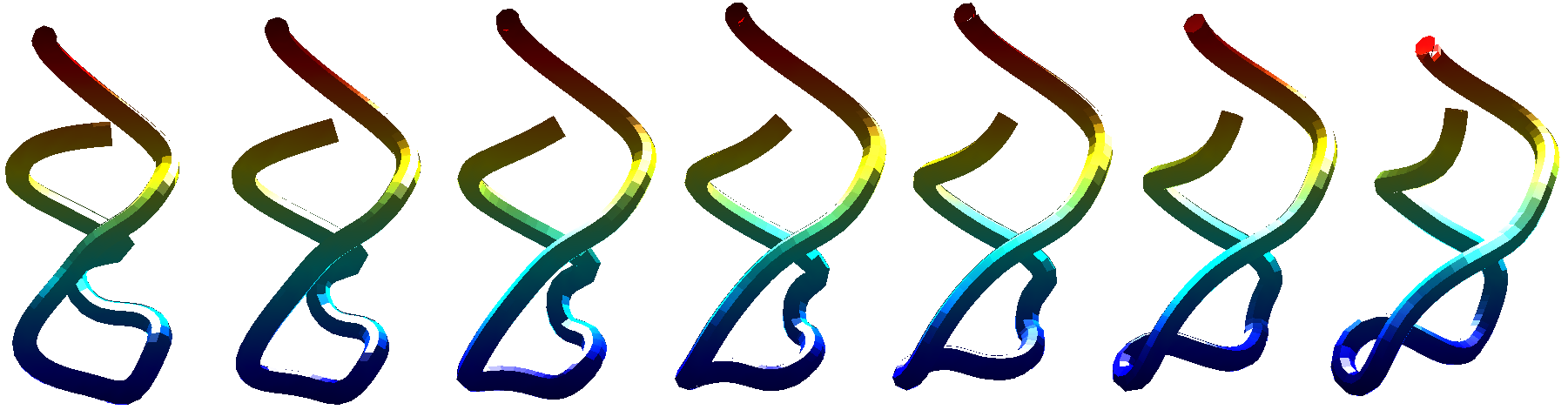
Here we propose an improved ESA framework that allows for a biologically relevant way to incorporate additional non-geometric information that tag the shape of the molecules (namely, the sequence of nucleotide/amino-acid letters for RNA/proteins and, in the latter case, also the labels for the so-called secondary structure). The biological representation is chosen such that the ESA framework continues to be mathematically formal. We have achieved superior classification rates compared to state-of-the-art methods on their RNA sets.
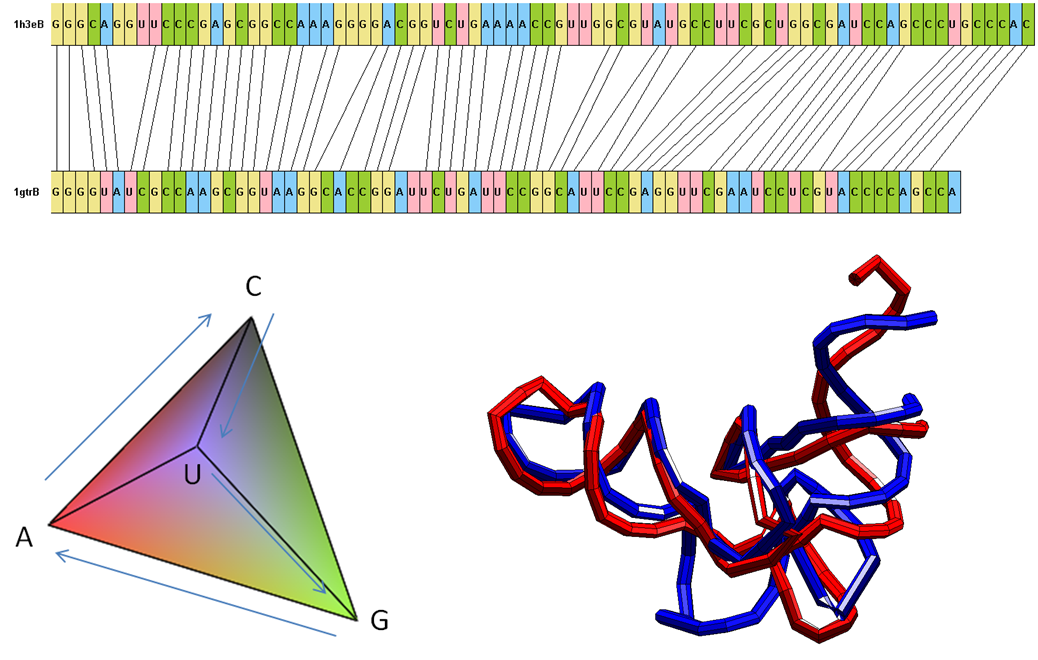
Based on the ESA distances, we have also developed a fast method to classify protein domains by the use of a set-reducing clustering-based technique we call Multiple Centroid Class Partitioning (MCCP). Comparison with other standard approaches for set reduction and classification show that MCCP significantly improves the accuracy while keeping the reduced sets smaller than the other methods.
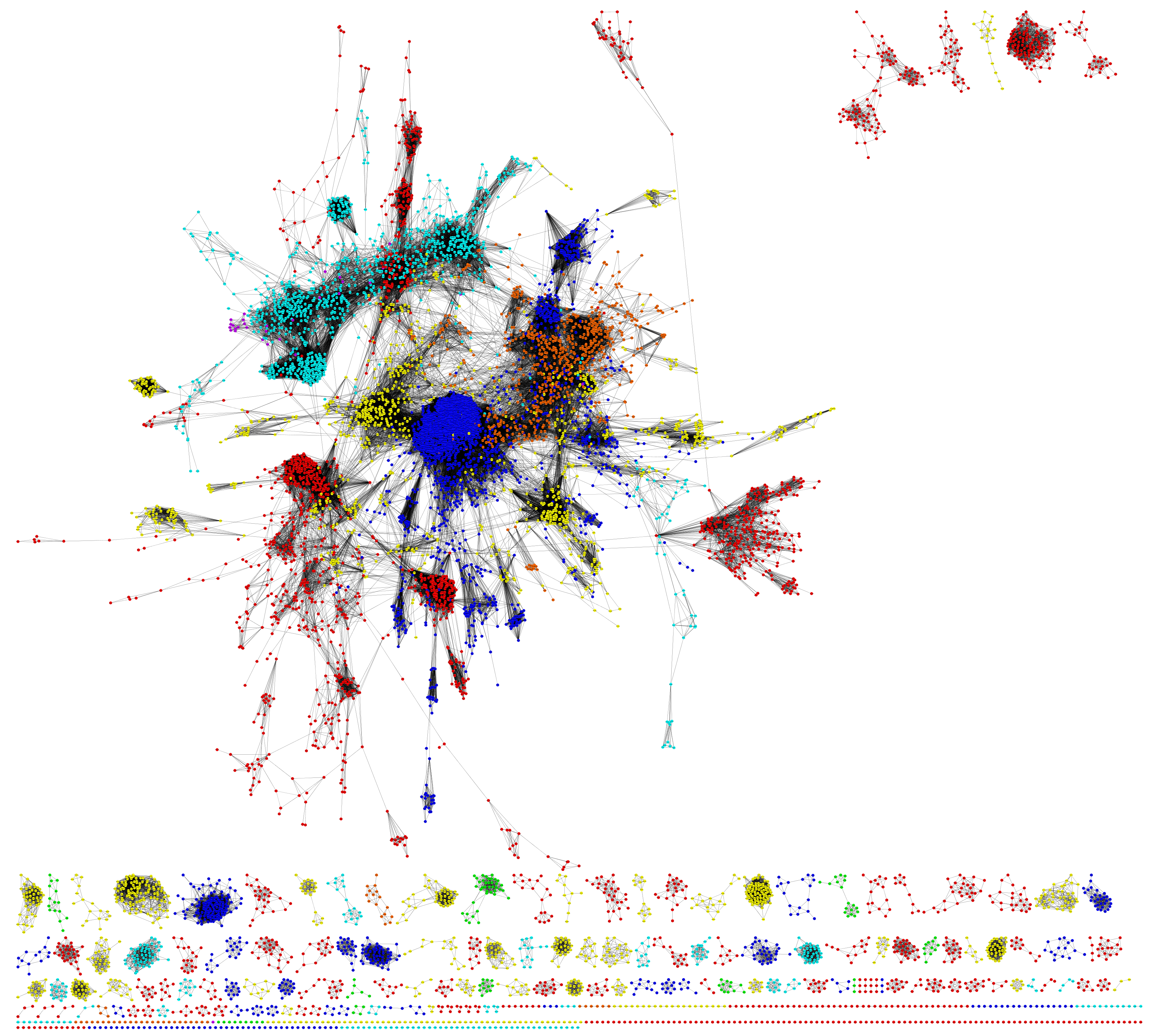
By proposing a network format derived from the distribution of ESA distances, this work also explores the plausibility of challenging current tree-like classification schemes for protein structures which are currently accepted as gold standards (such as SCOP or CATH). The literature agrees that the current gold standards are far from perfect and we have found more evidence to support this.
Please visit our research group web page:
Statistical Shape and Modeling Group
